GEN220_2020
2020 Edition of the Class
Project maintained by biodataprog Hosted on GitHub Pages — Theme by mattgraham
Finding Orthologs and Paralogs
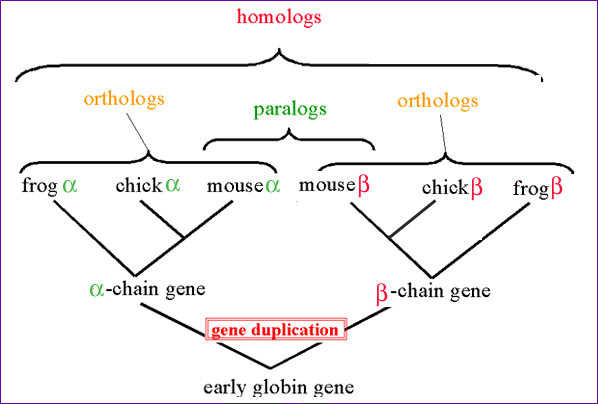
Gene families and Orthology
Problem: How to find “same” genes across multiple species.
Genes can duplicate (Paralogs) and can be identical due to descent (Ortholog)
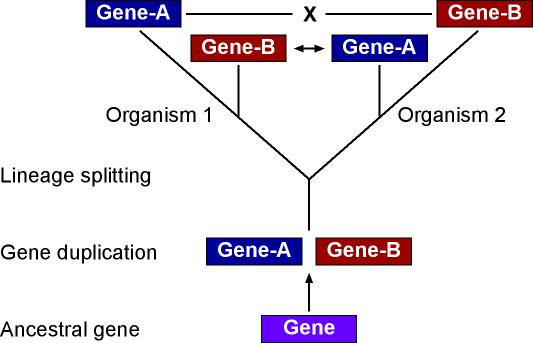
Methods
- BLAST: 1 way BLAST (Gene A in Species X, what is best hit in Species Y)
- BLAST: reciprocal BLAST
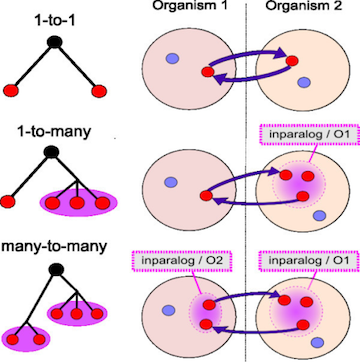
Trees can help resolve relationships
Best hits can sometimes be wrong (B) though it can be resolved with phylogenetics.
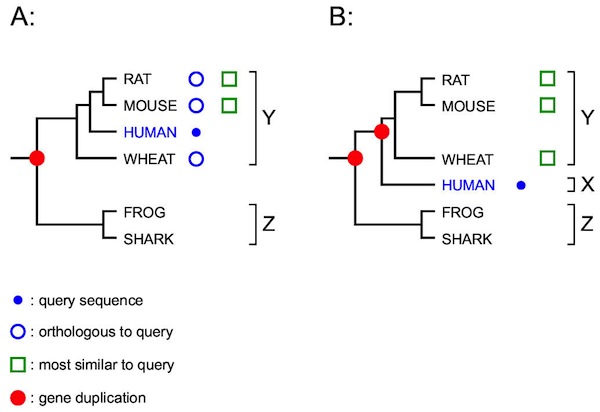
Reciprocal Searches
- Bi-directional or Reciprocal BLAST
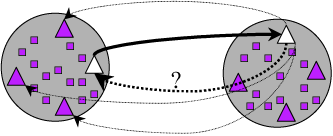
Implement Bidirectional
Method to find best top hit in one direction and the reverse.
Let’s walk through the code
Will write this in Python in Class
Clustering
- Lumping genes together based on similarity linkage
- Single-linkage means if there is a link between A-B then they are in a cluster
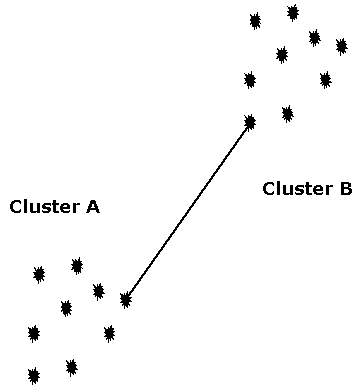
Code up single-linkage
Let’s look at some code.
Will write this in Python in Class
Issues
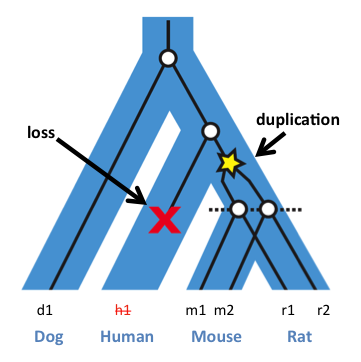
Tools to go after Orthologous and Paralogous equences
Steps to build orthologs on cluster
We will take 3 datasets of annotated Cyanobacteria, download and run analysis to generate Ortholog table.
#!/usr/bin/bash
#SBATCH --ntasks 16 --mem 8G -p short
module load ncbi-blast
module load orthofinder
module load miniconda2
CPU=8
mkdir -p cyanobacteria
cd cyanobacteria
curl -L -O ftp://ftp.ensemblgenomes.org/pub/bacteria/release-45/fasta/bacteria_10_collection/oscillatoriales_cyanobacterium_jsc_12/pep/Oscillatoriales_cyanobacterium_jsc_12.ASM30994v1.pep.all.fa.gz
curl -L -O ftp://ftp.ensemblgenomes.org/pub/bacteria/release-45/fasta/bacteria_0_collection/nostoc_punctiforme_pcc_73102/pep/Nostoc_punctiforme_pcc_73102.ASM2002v1.pep.all.fa.gz
curl -L -O ftp://ftp.ensemblgenomes.org/pub/bacteria/release-45/fasta/bacteria_4_collection/cyanobacterium_aponinum_pcc_10605/pep/Cyanobacterium_aponinum_pcc_10605.ASM31767v1.pep.all.fa.gz
# uncompress files and name them all *.fasta
for file in *.fa.gz
do
m=$(basename $file .pep.all.fa.gz)
pigz -dc $file > $m.fasta
done
cd ..
orthofinder.py -a $CPU -f cyanobacteria
Ortholog results
The output file by default will be the date of the anlaysis. Opening the file cyanobacteria/Results_XXX/Orthogroups.txt but I made a folder in the examples you look over. Here’s one table
Format
GroupName\tSp1_Gene1, Sp1_Gene2\tSp2_Gene1, Sp2_Gene2\tSp3_Gene1, Sp3_Gene2
Cyanobacterium_aponinum_pcc_10605.ASM31767v1 Nostoc_punctiforme_pcc_73102.ASM2002v1 Oscillatoriales_cyanobacterium_jsc_12.ASM30994v1
OG0000000 EKQ66605, EKQ66611, EKQ66662, EKQ66782, EKQ66954, EKQ66984, EKQ67084, EKQ67085, EKQ67278, EKQ67284, EKQ67330, EKQ67344, EKQ67361, EKQ67366, EKQ67368, EKQ67427,
EKQ67433, EKQ67590, EKQ67680, EKQ67799, EKQ67807, EKQ67983, EKQ68026, EKQ68032, EKQ68054, EKQ68173, EKQ68296, EKQ68354, EKQ68495, EKQ68500, EKQ68912, EKQ69012, EKQ69027, EKQ69073, EKQ69085,
EKQ69279, EKQ69300, EKQ69345, EKQ69368, EKQ69506, EKQ69549, EKQ69629, EKQ69630, EKQ69655, EKQ69739, EKQ69760, EKQ69872, EKQ69984, EKQ70005, EKQ70023, EKQ70046, EKQ70130, EKQ70179, EKQ70602, E
KQ70786, EKQ70840, EKQ70870, EKQ70894, EKQ71088, EKQ71090, EKQ71265, EKQ71335
OG0000001 AFZ52442, AFZ54265, AFZ54640 ACC78968, ACC78978, ACC79054, ACC79090, ACC79138, ACC79182, ACC79941, ACC80158, ACC80645, ACC81000, ACC81451, ACC81520, ACC81612, ACC81781, ACC
81797, ACC82091, ACC82628, ACC82978, ACC83035, ACC83215, ACC83711, ACC84528, ACC84844, ACC84851, ACC85118, ACC85266, ACC85421 EKQ66618, EKQ66620, EKQ66621, EKQ67508, EKQ68220, EKQ69508, EKQ
69971, EKQ69995, EKQ70003, EKQ70556, EKQ70833, EKQ71286
OG0000002 AFZ55137 ACC79344, ACC80485, ACC80595, ACC82143, ACC82836, ACC82962, ACC83845, ACC83999, ACC84046, ACC84047, ACC84257, ACC84700, ACC84955, ACC84966, ACC84970, ACC84971,
ACC84972, ACC84974, ACC84981, ACC84982, ACC84983, ACC85032 EKQ66950, EKQ67597, EKQ67615, EKQ68095, EKQ68496, EKQ68499, EKQ69362, EKQ69910, EKQ69959
OG0000003 AFZ53198 ACC78875, ACC78976, ACC79256, ACC79524, ACC79759, ACC80145, ACC80528, ACC80591, ACC80631, ACC80670, ACC80818, ACC81317, ACC81502, ACC82310, ACC82674, ACC82718,
ACC82769, ACC83025, ACC83081, ACC83457, ACC83602, ACC83721, ACC83749, ACC84422, ACC85331 EKQ69908
OG0000004 ACC80422, ACC80525, ACC80662, ACC80851, ACC80857, ACC80914, ACC81440, ACC81547, ACC81570, ACC81705, ACC81945, ACC81949, ACC81968, ACC81987, ACC82586, ACC83096, ACC8342
6, ACC83981, ACC84622, ACC84732, ACC85457 EKQ66830, EKQ66911, EKQ67039, EKQ67311, EKQ69997, EKQ70735
OG0000005 AFZ52318, AFZ52611, AFZ52613, AFZ52925, AFZ52973, AFZ53626, AFZ53840, AFZ53841, AFZ53849, AFZ53949, AFZ54004, AFZ54551, AFZ54904, AFZ55229 ACC80197, ACC81960, ACC82068, A
CC82559, ACC83603, ACC83674, ACC85005, ACC85009 EKQ67574, EKQ67809, EKQ69976
OG0000006 AFZ52319, AFZ53394, AFZ54017, AFZ54472 ACC79360, ACC79745, ACC79853, ACC80832, ACC80867, ACC81560, ACC82363, ACC82406, ACC82436, ACC82585, ACC84067, ACC84385 EKQ67267, EKQ67
478, EKQ67551, EKQ67724, EKQ67810, EKQ68266
OG0000007 AFZ53704, AFZ54461, AFZ54462 ACC79786, ACC80242, ACC80282, ACC80538, ACC80768, ACC81387, ACC82067, ACC83125, ACC83317, ACC83447, ACC84291, ACC84302, ACC85008 EKQ6767
2, EKQ68369, EKQ70142, EKQ70145, EKQ71300
The tool also generates summary statistics we can look through.
Could write a script to turn this into a table or use the summary count table provided.
ORTHOLOG_GRP SP1 SP2 SP3
ORTHO_0001 10 5
ORTHO_0002 1 1