RNASeq
Efforts to sequence the transcripts expressed in a cell or organism.
Statistics
Measuring gene expression
Resources
- See the StatQuest Video on RPKM/FPKM/TPM to better understand how the statistics can be used to evaluate gene expression.
Using techniques to extract the
Wang et al. Nat Rev Genetics. 2009. doi:10.1038/nrg2484
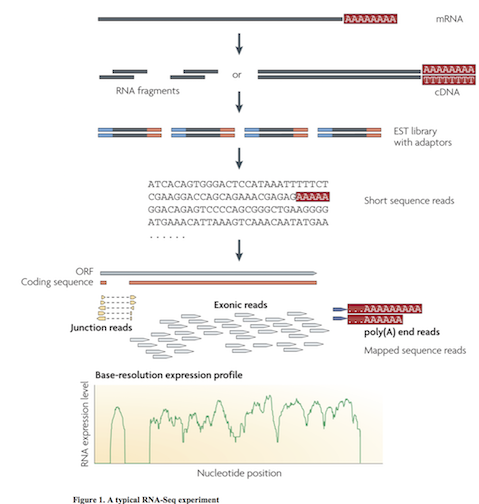
Multiple approaches to understanding the transcriptome
- Genome sequenced, align RNAseq reads to genome
-
de novo Assembly of mRNA into transcripts
- Quantify gene expression from reads aligned to genome or transcripts
Reads to Genome mapping
It is important to note that aligning sequences to the genome when there are introns requires dealing with introns. So splice-aware alignements are needed in some cases.
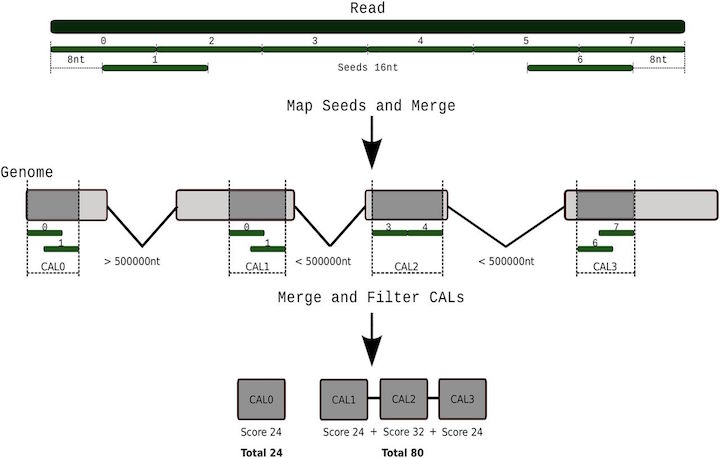
Tarraga et al 2017. DNA Research.10.1093/dnares/dsv039
#Reads to Genome mapping
Challenges: mRNA is spliced, genome contains introns
Splice-aware short read aligners. Speed and accuracy tradeoffs
- Tophat + Bowtie – this is old don’t use
- HISAT2
- GMAP/GSNAP
- STAR
Need to Quantify expression
- Count reads overlapping exons
- Table of total read counts per gene
- Normalize counts for gene length and sequencing library depth
- Gene expression then is FPKM - Fragments per Kilobase per Millions of reads
- Tools: htseq-count, stringtie
- SubRead
- BEDtools
- R tools with iRanges
Evaluating expression differences
Statistical tools for evaluating gene expression differences
- Ballgown bioconductor package
- DESeq bioconductor package
- edgeR bioconductor package
Alternative approach for Quantifying
Compare reads to Transcripts instead of Genome
- Kalisto and Sailfish are common tools
- Bray et al 2016 “Near-optimal probabilistic RNA-seq quantification” doi:10.1038/nbt.3519
- Patro et al 2014 “Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms” doi:10.1038/nbt.2862
Alignment free quantification
Usage: kallisto quant [arguments] FASTQ-files
Required arguments:
-i, --index=STRING Filename for the kallisto index to be used for
quantification
-o, --output-dir=STRING Directory to write output to
Optional arguments:
--bias Perform sequence based bias correction
-b, --bootstrap-samples=INT Number of bootstrap samples (default: 0)
--seed=INT Seed for the bootstrap sampling (default: 42)
--plaintext Output plaintext instead of HDF5
--fusion Search for fusions for Pizzly
--single Quantify single-end reads
--single-overhang Include reads where unobserved rest of fragment is
predicted to lie outside a transcript
--fr-stranded Strand specific reads, first read forward
--rf-stranded Strand specific reads, first read reverse
-l, --fragment-length=DOUBLE Estimated average fragment length
-s, --sd=DOUBLE Estimated standard deviation of fragment length
(default: -l, -s values are estimated from paired
end data, but are required when using --single)
-t, --threads=INT Number of threads to use (default: 1)
--pseudobam Save pseudoalignments to transcriptome to BAM file
--genomebam Project pseudoalignments to genome sorted BAM file
-g, --gtf GTF file for transcriptome information
(required for --genomebam)
-c, --chromosomes Tab separated file with chromosome names and lengths
(optional for --genomebam, but recommended)
Note this won’t quite work to copy and paste.
#!/usr/bin/bash
module load kallisto
ln -s /bigdata/gen220/shared/data-examples/rnaseq/kallisto/S_cerevisiae_ORFs.fasta
ln -s
kallisto index -i Scer.idx S_cerevisiae_ORFs.fasta
cat samples.tsv | while read ACC COND REP
do
OUT=output/$COND.$REP
kallisto quant -t 8 --single -l 300 -s 20 -i Scer.idx -o $OUT data/${ACC}_1.fastq.gz
done
Go see /bigdata/gen220/shared/data-examples/rnaseq/kallisto
Denovo assembly
Trinity Assembler for RNASeq
$ module load trinity-rnaseq
$ module switch perl/5.22.0
$ Trinity --seqType fq --left reads_1.fq --right reads_2.fq --CPU 8 --max_memory 20G
ORF identification
Once we have assembled the transcriptome, want to find genes in there.
- Finds Open Reading Frames in mRNA transcripts
$ module load transdecoder
$ TransDecoder.LongOrfs -t target_transcripts.fasta
RNAseq read mapping
Using HISAT2 for RNAseq read mapping
Download those files.
# start an interactive session
srun -N 1 -n 4 -p short --mem 16gb --pty bash -l
module load hisat2
# uncompress
gunzip S_cerevisiae.gff3.gz S_cerevisiae.fasta.gz
# build index
hisat2-build S_cerevisiae.fasta yeast
# run search
ln -s /bigdata/gen220/shared/data-examples/rnaseq/yeast_rnaseq/*.gz .
hisat2 -x yeast -1 SRR3396381_1.fastq.gz -2 SRR3396381_2.fastq.gz -S SRR3396381.sam -p 4
module load samtools
samtools view -Ob -o SRR3396381.bam SRR3396381.sam
samtools sort -o SRR3396381.sort.bam SRR3396381.bam
samtools index SRR3396381.sort.bam SRR3396381.bam
samtools flagstat SRR3396381.sort.bam
Get counts
Subread - http://subread.sourceforge.net/
module load subread
GENOME=S_cerevisae.fasta
GFF=S_cerevisae.gff3
OUTFILE=SRR3396381.tab
INFILE=SRR3396381.sort.bam
featureCounts -g gene_id -T $CPUS -G $GENOME -s 0 -a $GFF -o $OUTFILE \
-F GTF $INFILE
Template for Projects
Here’s a template for RNASeq analyses
https://github.com/biodataprog/RNASeq_template Click on ‘Use this template’ - you can create your own version of this.
It will prompt you give it a name.
Go to the command line to download.
git clone yourname/YourRNASeqAnalysis.git
Edit samples.csv to describe names of some experiments SRR3396381.
Download data in input folder.
Download or us download script to get genome files (need to put a genome FASTA file in the folder). If want to do kallisto will need a mRNA file of transcriptome.